[TOC]
人体3D建模:
SMPL 2015 《A Skinned Multi-Person Linear Model》
Loper, Matthew and Mahmood, Naureen and Romero, Javier and Pons-Moll, Gerard and Black, Michael J.
Official https://smpl.is.tue.mpg.de/
SMPL https://github.com/CalciferZh/SMPL
Numpy, TF and PyTorch implementation of human body SMPL model and infant body SMIL model.
参数化的人体3D模型生成工具
SMPLify 2016 论文:《Keep it SMPL: Automatic Estimation of 3D Human Pose and Shape from a Single Image》
官网:http://smplify.is.tue.mpg.de/
github: https://github.com/vchoutas/smplify-x
论文解读: https://eros-l.github.io/weekly/2018/11/28/weekly5/
项目实践: 官网数据:
smplify_code_v2.zip
lsp_results.tar.gz
human_eva_results.tar.gz
h36M_results_wtout_plytar.gz
常见问题:
—–自定义自己的数据集
使用经过LSP训练过的关节检测器CPM/DeepCut
fit_3d.py 的run_sigle_fit
Ubuntu下python3环境:
下载lsp数据集。
DeepCut 提取lsp数据集2D Pose特征点 (DeepCut 提取的 lsp dataset 的二维特征点)
SMPLify fit_3d.py 转换人体3D Pose的模型截图jpg和pkl格式。
pkl 文件来生成对应的 obj 文件或者其他格式的 mesh(未完成)
参考文档:
1.学习报告(week 5)
2.(week 2)【经验教训】如何将CPM和SMPL的输入替换成任意图片并得到人体三维模型
SMPL-H(mano) 2017 官网:https://mano.is.tue.mpg.de/
论文:《Embodied Hands: Modeling and Capturing Hands and Bodies Together》 2017
Javier Romero*, Dimitrios Tzionas* and Michael J Black
SIGGRAPH ASIA 2017, BANGKOK, THAILAND
SMPLX 2019 官网:https://smpl-x.is.tue.mpg.de/
github: https://github.com/vchoutas/smplx
《Expressive Body Capture: 3D Hands, Face, and Body from a Single Image》
G. Pavlakos* , V. Choutas* , N. Ghorbani , T. Bolkart , A. A. A. Osman , D. Tzionas and M. J. Black
CVPR 2019
增加了对手,脸部,身体的细节的完善
pytorch实现,摈弃了chumpy
SMPLify-X https://github.com/vchoutas/smplify-x
从单张图片的3D人体复原
AMASS dataset https://amass.is.tuebingen.mpg.de/
AMASS is a large database of human motion unifying different optical marker-based motion capture datasets by representing them within a common framework and parameterization. AMASS is readily useful for animation, visualization, and generating training data for deep learning.
AMASS是一个大型的人类运动数据库,通过在共同的框架和参数化中表示不同的基于光学标记的运动捕获数据集来统一这些数据。AMASS对于动画,可视化以及生成用于深度学习的训练数据非常有用。
SMPL源码下载:http://smpl.is.tue.mpg.de/downloads 需要注册
本文主要讨论SMPL源码核心代码。需要储备的知识还有:三维重建基础知识,以及 chumpy阅读 以及opendr阅读 ,LBS DQS 等
SMPL文件主要包括 vert.py serialization.py lbs.py。下面将逐一说说明。
0、关节位置
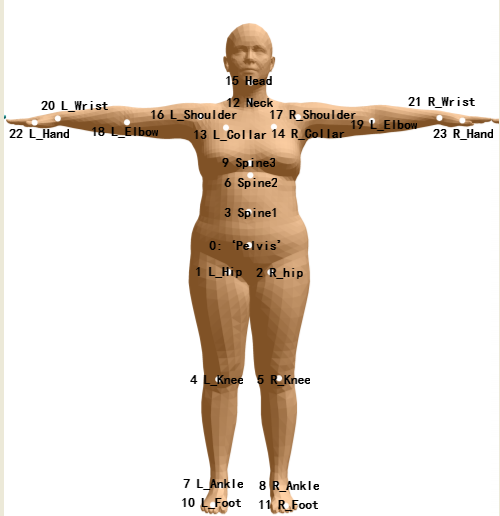
SMPL模型关节点名称
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 self.j_names = { 0: ‘Pelvis’, 1: ‘L_Hip’, 2: ‘R_Hip’, 3: ‘Spine1’, 4: ‘L_Knee’, 5: ‘R_Knee’, 6: ‘Spine2’, 7: ‘L_Ankle’, 8: ‘R_Ankle’, 9: ‘Spine3’, 10: ‘L_Foot’, 11: ‘R_Foot’, 12: ‘Neck’, 13: ‘L_Collar’, 14: ‘R_Collar’, 15: ‘Head’, 16: ‘L_Shoulder’, 17: ‘R_Shoulder’, 18: ‘L_Elbow’, 19: ‘R_Elbow’, 20: ‘L_Wrist’, 21: ‘R_Wrist’, 22: ‘L_Hand’, 23: ‘R_Hand’, }
关节树图形:
1、posemapper.py 主要是实现关节角度到姿势混合形状的映射。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 import chumpy as chimport numpy as npimport cv2class Rodrigues (ch.Ch): dterms = 'rt' def compute_r (self ): return cv2.Rodrigues(self .rt.r)[0 ] def compute_dr_wrt (self, wrt ): if wrt is self .rt: return cv2.Rodrigues(self .rt.r)[1 ].T def lrotmin (p ): if isinstance (p, np.ndarray): p = p.ravel()[3 :] return np.concatenate([(cv2.Rodrigues(np.array(pp))[0 ]-np.eye(3 )).ravel() for pp in p.reshape((-1 ,3 ))]).ravel() if p.ndim != 2 or p.shape[1 ] != 3 : p = p.reshape((-1 ,3 )) p = p[1 :] return ch.concatenate([(Rodrigues(pp)-ch.eye(3 )).ravel() for pp in p]).ravel() def posemap (s ): if s == 'lrotmin' : return lrotmin else : raise Exception('Unknown posemapping: %s' % (str (s),))
2、LBS.py 就是实现LBS
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 from posemapper import posemapimport chumpyimport numpy as npdef global_rigid_transformation (pose, J, kintree_table, xp ): results = {} pose = pose.reshape((-1 ,3 )) id_to_col = {kintree_table[1 ,i] : i for i in range (kintree_table.shape[1 ])} parent = {i : id_to_col[kintree_table[0 ,i]] for i in range (1 , kintree_table.shape[1 ])} if xp == chumpy: from posemapper import Rodrigues rodrigues = lambda x : Rodrigues(x) else : import cv2 rodrigues = lambda x : cv2.Rodrigues(x)[0 ] with_zeros = lambda x : xp.vstack((x, xp.array([[0.0 , 0.0 , 0.0 , 1.0 ]]))) results[0 ] = with_zeros(xp.hstack((rodrigues(pose[0 ,:]), J[0 ,:].reshape((3 ,1 ))))) for i in range (1 , kintree_table.shape[1 ]): results[i] = results[parent[i]].dot(with_zeros(xp.hstack(( rodrigues(pose[i,:]), ((J[i,:] - J[parent[i],:]).reshape((3 ,1 ))) )))) pack = lambda x : xp.hstack([np.zeros((4 , 3 )), x.reshape((4 ,1 ))]) results = [results[i] for i in sorted (results.keys())] results_global = results if True : results2 = [results[i] - (pack( results[i].dot(xp.concatenate( ( (J[i,:]), 0 ) ))) ) for i in range (len (results))] results = results2 result = xp.dstack(results) return result, results_global def verts_core (pose, v, J, weights, kintree_table, want_Jtr=False , xp=chumpy ): A, A_global = global_rigid_transformation(pose, J, kintree_table, xp) T = A.dot(weights.T) rest_shape_h = xp.vstack((v.T, np.ones((1 , v.shape[0 ])))) v =(T[:,0 ,:] * rest_shape_h[0 , :].reshape((1 , -1 )) + T[:,1 ,:] * rest_shape_h[1 , :].reshape((1 , -1 )) + T[:,2 ,:] * rest_shape_h[2 , :].reshape((1 , -1 )) + T[:,3 ,:] * rest_shape_h[3 , :].reshape((1 , -1 ))).T v = v[:,:3 ] if not want_Jtr: return v Jtr = xp.vstack([g[:3 ,3 ] for g in A_global]) return (v, Jtr)
3、vert.py verts_decorated函数没有被用到就不过多注释了
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 import chumpyimport lbsfrom posemapper import posemapimport scipy.sparse as spfrom chumpy.ch import MatVecMultdef ischumpy (x ): return hasattr (x, 'dterms' ) def verts_decorated (trans, pose, v_template, J, weights, kintree_table, bs_style, f, bs_type=None , posedirs=None , betas=None , shapedirs=None , want_Jtr=False ): for which in [trans, pose, v_template, weights, posedirs, betas, shapedirs]: if which is not None : assert ischumpy(which) v = v_template if shapedirs is not None : if betas is None : betas = chumpy.zeros(shapedirs.shape[-1 ]) v_shaped = v + shapedirs.dot(betas) else : v_shaped = v if posedirs is not None : v_posed = v_shaped + posedirs.dot(posemap(bs_type)(pose)) else : v_posed = v_shaped v = v_posed if sp.issparse(J): regressor = J J_tmpx = MatVecMult(regressor, v_shaped[:,0 ]) J_tmpy = MatVecMult(regressor, v_shaped[:,1 ]) J_tmpz = MatVecMult(regressor, v_shaped[:,2 ]) J = chumpy.vstack((J_tmpx, J_tmpy, J_tmpz)).T else : assert (ischumpy(J)) assert (bs_style=='lbs' ) result, Jtr = lbs.verts_core(pose, v, J, weights, kintree_table, want_Jtr=True , xp=chumpy) tr = trans.reshape((1 ,3 )) result = result + tr Jtr = Jtr + tr result.trans = trans result.f = f result.pose = pose result.v_template = v_template result.J = J result.weights = weights result.kintree_table = kintree_table result.bs_style = bs_style result.bs_type =bs_type if posedirs is not None : result.posedirs = posedirs result.v_posed = v_posed if shapedirs is not None : result.shapedirs = shapedirs result.betas = betas result.v_shaped = v_shaped if want_Jtr: result.J_transformed = Jtr return result def verts_core (pose, v, J, weights, kintree_table, bs_style, want_Jtr=False , xp=chumpy ): if xp == chumpy: assert (hasattr (pose, 'dterms' )) assert (hasattr (v, 'dterms' )) assert (hasattr (J, 'dterms' )) assert (hasattr (weights, 'dterms' )) assert (bs_style=='lbs' ) result = lbs.verts_core(pose, v, J, weights, kintree_table, want_Jtr, xp) return result
4、serialization.py SMPL模型的序列化函数。
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 def save_model (model, fname ): m0 = model trainer_dict = {'v_template' : np.asarray(m0.v_template),'J' : np.asarray(m0.J),'weights' : np.asarray(m0.weights),'kintree_table' : m0.kintree_table,'f' : m0.f, 'bs_type' : m0.bs_type, 'posedirs' : np.asarray(m0.posedirs)} if hasattr (model, 'J_regressor' ): trainer_dict['J_regressor' ] = m0.J_regressor if hasattr (model, 'J_regressor_prior' ): trainer_dict['J_regressor_prior' ] = m0.J_regressor_prior if hasattr (model, 'weights_prior' ): trainer_dict['weights_prior' ] = m0.weights_prior if hasattr (model, 'shapedirs' ): trainer_dict['shapedirs' ] = m0.shapedirs if hasattr (model, 'vert_sym_idxs' ): trainer_dict['vert_sym_idxs' ] = m0.vert_sym_idxs if hasattr (model, 'bs_style' ): trainer_dict['bs_style' ] = model.bs_style else : trainer_dict['bs_style' ] = 'lbs' pickle.dump(trainer_dict, open (fname, 'w' ), -1 ) def backwards_compatibility_replacements (dd ): if 'default_v' in dd: dd['v_template' ] = dd['default_v' ] del dd['default_v' ] if 'template_v' in dd: dd['v_template' ] = dd['template_v' ] del dd['template_v' ] if 'joint_regressor' in dd: dd['J_regressor' ] = dd['joint_regressor' ] del dd['joint_regressor' ] if 'blendshapes' in dd: dd['posedirs' ] = dd['blendshapes' ] del dd['blendshapes' ] if 'J' not in dd: dd['J' ] = dd['joints' ] del dd['joints' ] if 'bs_style' not in dd: dd['bs_style' ] = 'lbs' def ready_arguments (fname_or_dict ): if not isinstance (fname_or_dict, dict ): dd = pickle.load(open (fname_or_dict)) else : dd = fname_or_dict backwards_compatibility_replacements(dd) want_shapemodel = 'shapedirs' in dd nposeparms = dd['kintree_table' ].shape[1 ]*3 if 'trans' not in dd: dd['trans' ] = np.zeros(3 ) if 'pose' not in dd: dd['pose' ] = np.zeros(nposeparms) if 'shapedirs' in dd and 'betas' not in dd: dd['betas' ] = np.zeros(dd['shapedirs' ].shape[-1 ]) for s in ['v_template' , 'weights' , 'posedirs' , 'pose' , 'trans' , 'shapedirs' , 'betas' , 'J' ]: if (s in dd) and not hasattr (dd[s], 'dterms' ): dd[s] = ch.array(dd[s]) if want_shapemodel: dd['v_shaped' ] = dd['shapedirs' ].dot(dd['betas' ])+dd['v_template' ] v_shaped = dd['v_shaped' ] J_tmpx = MatVecMult(dd['J_regressor' ], v_shaped[:,0 ]) J_tmpy = MatVecMult(dd['J_regressor' ], v_shaped[:,1 ]) J_tmpz = MatVecMult(dd['J_regressor' ], v_shaped[:,2 ]) dd['J' ] = ch.vstack((J_tmpx, J_tmpy, J_tmpz)).T dd['v_posed' ] = v_shaped + dd['posedirs' ].dot(posemap(dd['bs_type' ])(dd['pose' ])) else : dd['v_posed' ] = dd['v_template' ] + dd['posedirs' ].dot(posemap(dd['bs_type' ])(dd['pose' ])) return dd def load_model (fname_or_dict ): dd = ready_arguments(fname_or_dict) args = { 'pose' : dd['pose' ], 'v' : dd['v_posed' ], 'J' : dd['J' ], 'weights' : dd['weights' ], 'kintree_table' : dd['kintree_table' ], 'xp' : ch, 'want_Jtr' : True , 'bs_style' : dd['bs_style' ] } result, Jtr = verts_core(**args) result = result + dd['trans' ].reshape((1 ,3 )) result.J_transformed = Jtr + dd['trans' ].reshape((1 ,3 )) for k, v in dd.items(): setattr (result, k, v) return result
环境配置(SMPL) chumpy安装
pip install chumpy (0.86的chumpy已经支持python3 不需要特别修改代码安装)
python2和python3一些库的名字不同:
例如:no module named cPickle
pickle模块,在ython3中为import pickle,python2中为import cPickle as pickle
opendr 安装
https://codeload.github.com/polmorenoc/opendr/zip/master
1 python setup.py build
2 python setup.py install
OPENGL-python 安装
https://files.pythonhosted.org/packages/d7/8a/5db9096aa6506e405309c400bd0feb41997689cbba30683479c30dba6355/PyOpenGL-3.1.4.tar.gz
1- 解压
2- python setup.py build
3- python setup.py install
环境配置Ubuntu(SMPL) opendr
Mesa 3D Graphics Library
https://www.mesa3d.org/osmesa.html
1 2 sudo apt-get install libosmesa6-dev pip3 install opendr
PyOpenGL
1 pip3 install PyOpenGL PyOpenGL_accelerate
c参考资料: SMPL模型改用python3+numpy计算
SMPL: A Skinned Multi-Person Linear Model
人体模型介绍 - SMPL
3D相关学术人员及Blog 浙大博士 https://52zju.cn/?paged=2 三维重建